
In 2014, Google acquired a little-known artificial intelligence (AI) research company. It paid around $600 million.
It was considered a large sum at the time… especially for a startup that had only been around for a few years, had no real products, and very little revenue.
It wasn’t even clear what Google was going to do with the business. Or how it would monetize the work coming out of — what was essentially — a research laboratory.
In its early years, the team at DeepMind was focused on developing artificial intelligences capable of mastering complex games. It wasn’t because they expected to monetize the work. It was because they were complex challenges to solve.
The game of Go is a perfect example…
There are 10170 possible board configurations, making it impossible for any supercomputer to calculate every possible outcome. There just isn’t enough computing power on Earth to do so.
Which meant that DeepMind’s AlphaGo AI had to excel at pattern recognition. It actually had to “think” and determine the best strategy with each move to achieve the desired outcome — a win.
The results were stunning. By 2015, AlphaGo destroyed the world’s champion in Go, 4 games to 1, in a competition held in Seoul, Korea. It was a feat previously thought impossible given the complexity of the game, and yet it happened at least a decade before the “experts” thought possible.
And while the news outlets were focused on AlphaGo’s win, much in the same way that they were in 1997 when IBM’s Deep Blue beat chess champion Kasparov, there was something much bigger happening at DeepMind.
Back then, the DeepMind team was in the earlier stages of developing artificial intelligence capable of solving one of the grand challenges of life sciences — how proteins fold.
As we learned earlier this week in Outer Limits — The $1 Billion Biotech Idea, proteins are constituted by a long string of amino acids, which determine the ultimate structure of the protein itself — or how proteins fold.
Proteins are the building blocks of life, and understanding their structure enables us to understand their functions, as well as how to optimize therapies to improve health and save lives.
Understanding the structure of proteins has long been a grand challenge… with limited success for solving using physics-based modeling.
The problem was so important, every year since 1994, an experiment known as the Critical Assessment of Structure Prediction (CASP) — let’s call it a competition — was run.
The goal of CASP was to see which teams most accurately predicted protein structures.
Here’s the basis of the competition…
Competitors are given information about a protein. This includes sequences of amino acids, for example. But the structure of the protein is unknown.
Using computational methods, algorithms, and physics-based modeling, contenders attempt to predict how the protein folds into a 3D shape. They try to figure out the arrangement of the atoms in the protein.
After the teams submit their predictions to CASP organizers, the predictions are compared to the actual experimental structures — which had been kept secret.
CASP organizers reveal the results, including the accuracy of the prediction, how refined it was, and where contenders went right or wrong. This feedback helps with the improvement and refinement of methods and modeling for future predictions.
But things got really interesting when AI came into the picture.
2018 was the year in which DeepMind unleashed its AlphaFold AI on the CASP challenge.
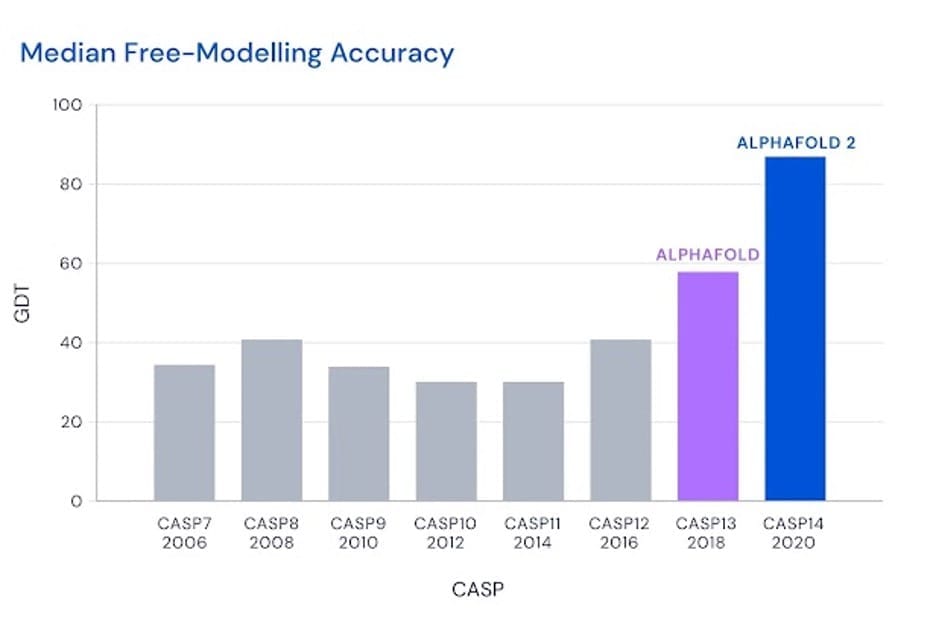
It was head and shoulders above the best-in-class from prior years, approaching 60 GDT.
GDT stands for Global Distance Test, a representative of how far off the protein structure predictions are from the known correct position.
A GDT of 60 wasn’t very useful from the perspective of accuracy and ultimately drug development. We can think of a GDT score very loosely as the percentage of being 100% accurate. 60 is not very accurate, in other words — nowhere near being accurate enough for drug development.
But by 2020, that number jumped to a median GDT score of 87. That was an incredible moment, and something I covered in detail in my former publication, The Bleeding Edge.
Again, it was still not close enough in terms of accuracy, but the jump in improvement gave promise and hope that the team at DeepMind would be able to crack the code of life.
The incredible breakthrough came in the summer of 2022, when DeepMind released an updated version of its AlphaFold AI. It came with a database — free for all to see — of protein structure predictions of more than 200 million known proteins.
Better yet, the GDT score of the new AlphaFold (AlphaFold2) was an incredible 92.4 (best possible score is 100).
In other words, AlphaFold2 now “predicted” more than 200 million known protein structures… it did so with a near-perfect prediction-to-actual GDT score.
It goes without saying, this feat would be impossible otherwise. Certainly far and beyond the capabilities of the human teams using physics-based modeling in the CASP competitions.
To visualize what a remarkable achievement this was, the images below show how greatly the knowledge base of proteins expanded… with just this one release of a single AI tool.
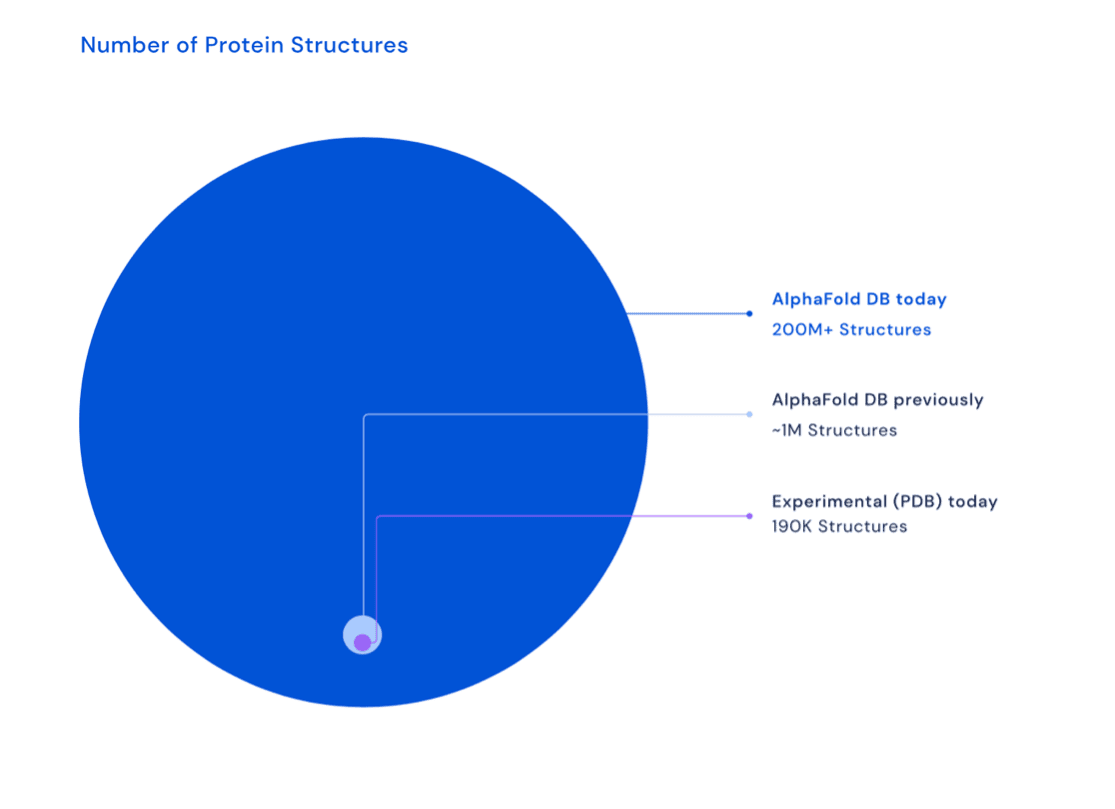
DeepMind went from about 1 million predicted protein structures in 2020, to more than 200 million protein structures in 2022.
And get this…
It wasn’t just for human protein structures.
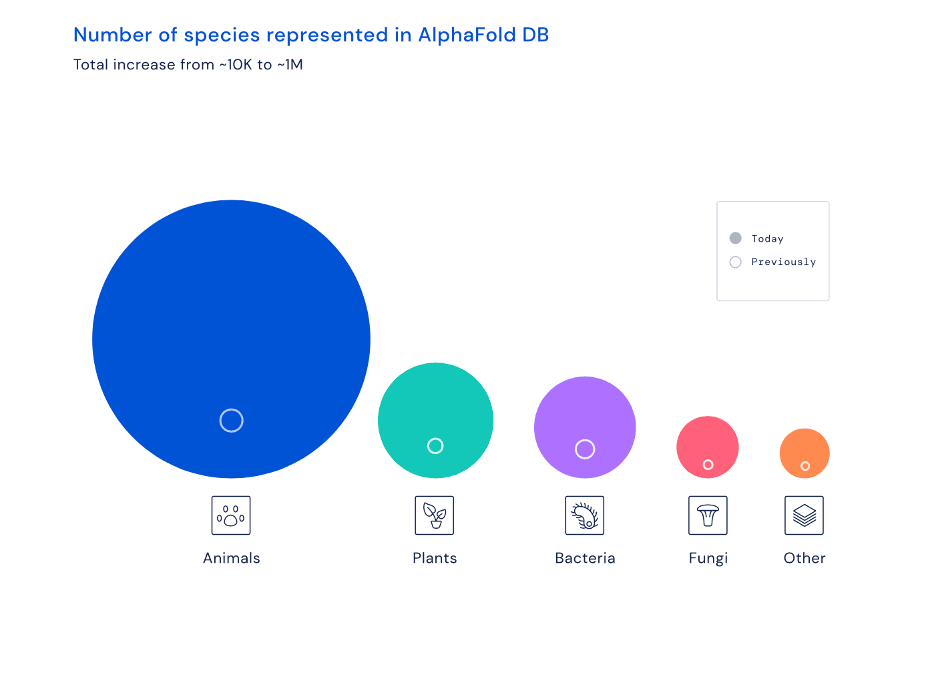
The 2022 DeepMind AlphaFold2 release also mapped out the protein structures of animals, plants, bacteria, and fungi.
The jump was from 10,000 to more than 1 million for non-human proteins.
The ramifications of this breakthrough were, and still are, awesome… even two years later.
Millions of researchers have been using this database in the time since, to do things like:
It almost felt like that was “it.” Problem solved.
Until yesterday.
Just yesterday, DeepMind released AlphaFold 3.
But it’s not what we might think.
It’s not some incremental improvement in accuracy, inching closer to a GDT score of 100.
AlphaFold 3 has predicted the structure — not just protein structures — of all molecules of life.
AlphaFold 3 is capable of accurately predicting the structures of proteins, DNA, RNA, and ligands — “binding” molecules that create bonds of various strengths with other molecules and ions. It even predicts how they interact.
This is likely the most valuable scientific tool and repository of life sciences information that the biotech could have ever asked for. And it’s free.
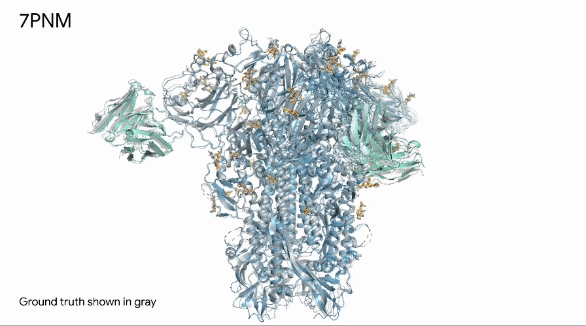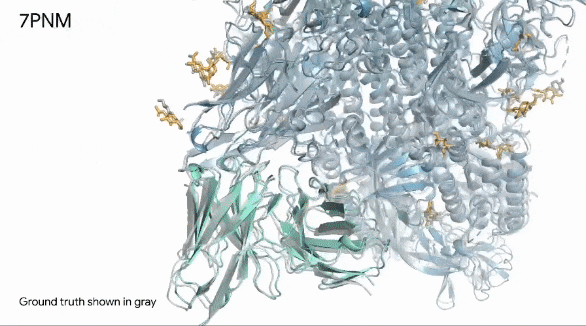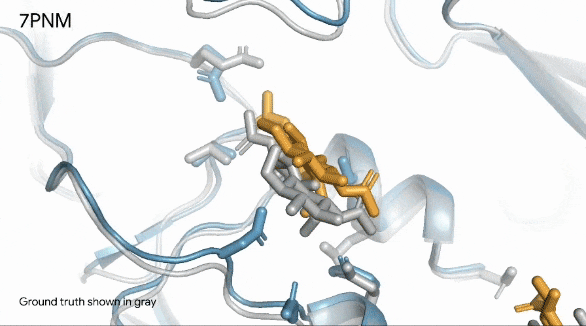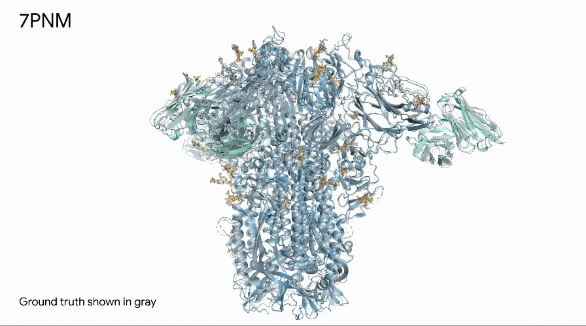
Below, we can see in remarkable clarity the spike protein (in blue) of a common cold virus. We can also see how human antibodies (in turquoise) — which is what our immune system produces to fight a virus — interact with the cold’s spike proteins.
They gray structure, seen in the image below, is the actual structure of the spike protein, antibodies, and their interaction.
The image shows how the prediction is exactly like the real world.
AlphaFold has been so widely used that it has already been cited more than 20,000 times in scientific research. Its existence has transformed life science research and the biotech industry, and this is only since 2022.
To build upon Outer Limits — Open Sourcing CRISPR and Profluent Bio’s release of OpenCRISPR, AlphaFold is now able to predict how a protein binds to DNA.
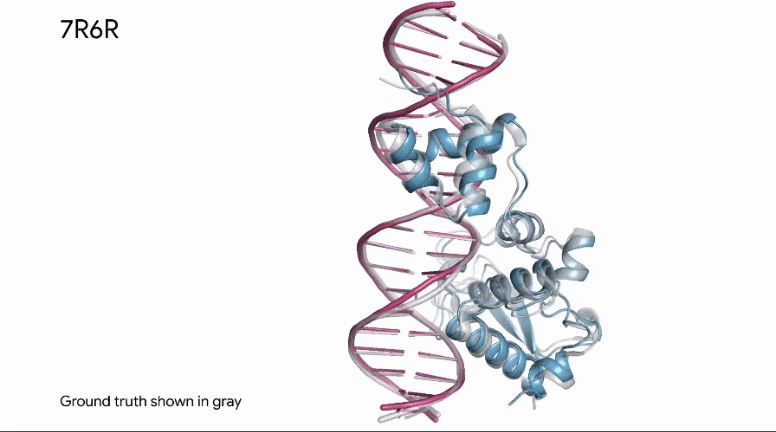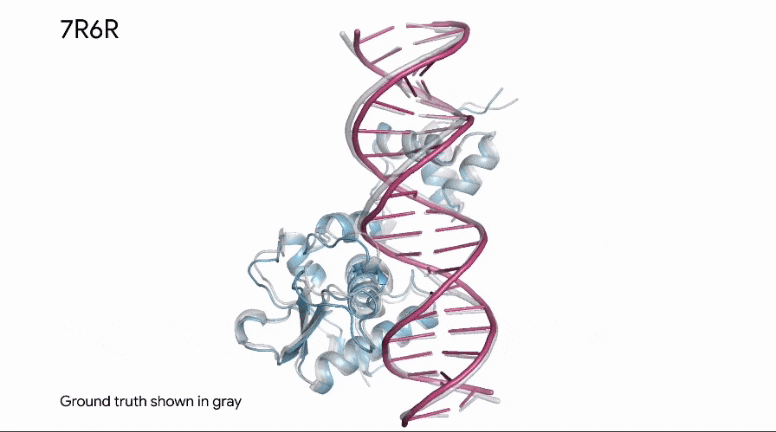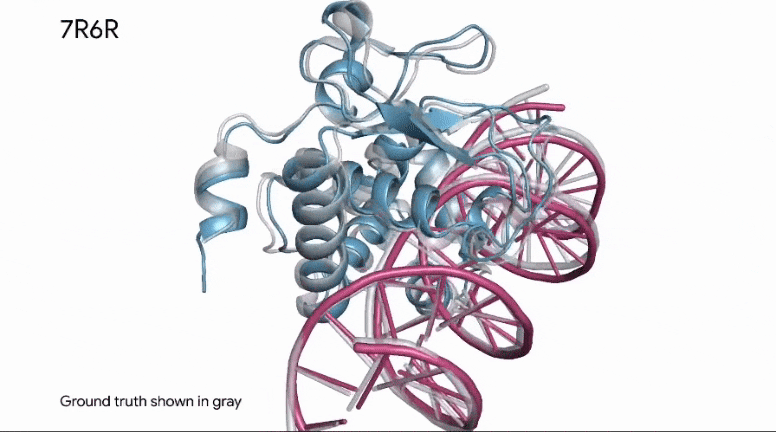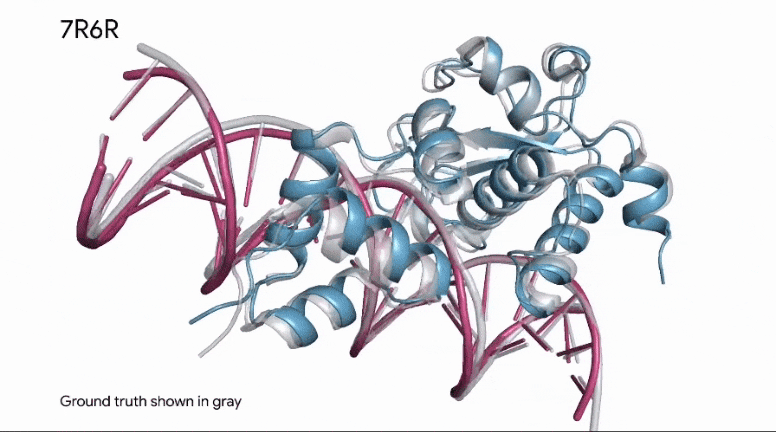
This kind of information is invaluable in research, as it applies to how known enzymes (proteins) will bind to DNA in genetic editing applications.
The same predictive capabilities can be applied in the analysis of synthetic proteins, as well.
We could argue that this information that DeepMind has provided the world is invaluable, perhaps priceless.
Which raises an important question…
Why give it away?
DeepMind has generated very limited revenues, historically, mostly from partnerships — outsourcing AI-related work for other companies and organizations.
After Google acquired DeepMind, it was incurring massive losses on the order of 470 million pounds in 2018, and 477 million pounds in 2019. (DeepMind is based in London.)
It was reported in 2021 — when DeepMind posted a profit as a division of Google — but that was only from an accounting perspective. DeepMind’s only customers were other divisions of Google, so these were just accounting cross-charges for services.
Over the years, Alphabet (Google) has spent more than $2 billion funding DeepMind’s research efforts.
I like to think of these grand gestures of open sourcing such incredible research as a way to pay penance (sarcasm) for Alphabet’s underlying business model, which is appropriating personal data from every user of Google products and selling access to that data to advertisers.
It became a $2 trillion company taking users’ data — in most cases without explicit consent and/or understanding — and now sits on more than $100 billion in cash. It can afford to spend a small portion of that on research as a way to pay back the world for its misdoings (again, sarcasm).
These kinds of gestures are, however, useful in the context of companies that have dominant positions in their industries and are under regulatory scrutiny. Google’s Android operating system has more than 70% of global market share on mobile phones. And Google’s search engine has more than 90% global market share.
And over the years, Google has had to pay billions in consumer privacy lawsuits where Google knowingly violated consumers’ privacy rights, secretly tracked consumers internet activity, and illegally tracked consumers physical locations.
Having a great story to tell regulators like AlphaFold is a smart counter to its business practice and abuse of power in the tech industry. The corporate positioning is that Alphabet’s not just a company for profit, but a company for the betterment of humanity. It’s nonsense, but easy to understand how lawyers would argue as such.
But what most don’t know is that in late 2021, Google spun out a new for-profit company — Isomorphic Laboratories — dedicated entirely to drug discovery.
Given Isomorphic’s incredible backing, financial resources, and AI talent out of DeepMind, it has been one of the most important biotech companies to watch. It was clear that the company would have a major impact on the industry.
So it wasn’t a surprise when, this January, it signed two major pharmaceutical partnerships with Eli Lilly and Novartis. The total value was $3 billion.
The primary goal of Isomorphic in these partnerships is to help the pharmaceutical giants reduce the time for drug discovery from an average of five years down to just two years.
The idea is simple. Using AI, Isomorphic will be able to present a drug for clinical trials within two years, instead of five.
That will result in less than half the time, and a fraction of the cost. And AI has the added benefit of presenting higher probability drug candidates for each drug discovery. And it will also be used to avoid ever sending low probability candidates through Phase 1 clinical trials.
This all means that drug discovery, clinical trials, and ultimately regulatory approvals will happen years faster than they have in the past. And it is highly likely that safety and efficacy will improve.
And of course, Alphabet (Google) will ultimately receive an incredible return on its investment in DeepMind.
The other stark reality is that Alphabet (Google) wants as many internet and mobile users as possible using its technology… so that it can track them and collect data.
The longer we all live, the more data to collect and sell, which means more advertising revenues.
And that is the real business of Alphabet (Google).
We always welcome your feedback. We read every email and address the most common comments and questions in the Friday AMA. Please write to us here.